First, please make sure that you have previously performed the pre-processing, functional annotation, gene regulatory network construction steps, see Pre-processing, Functional annotation, Gene regulatory network construction.
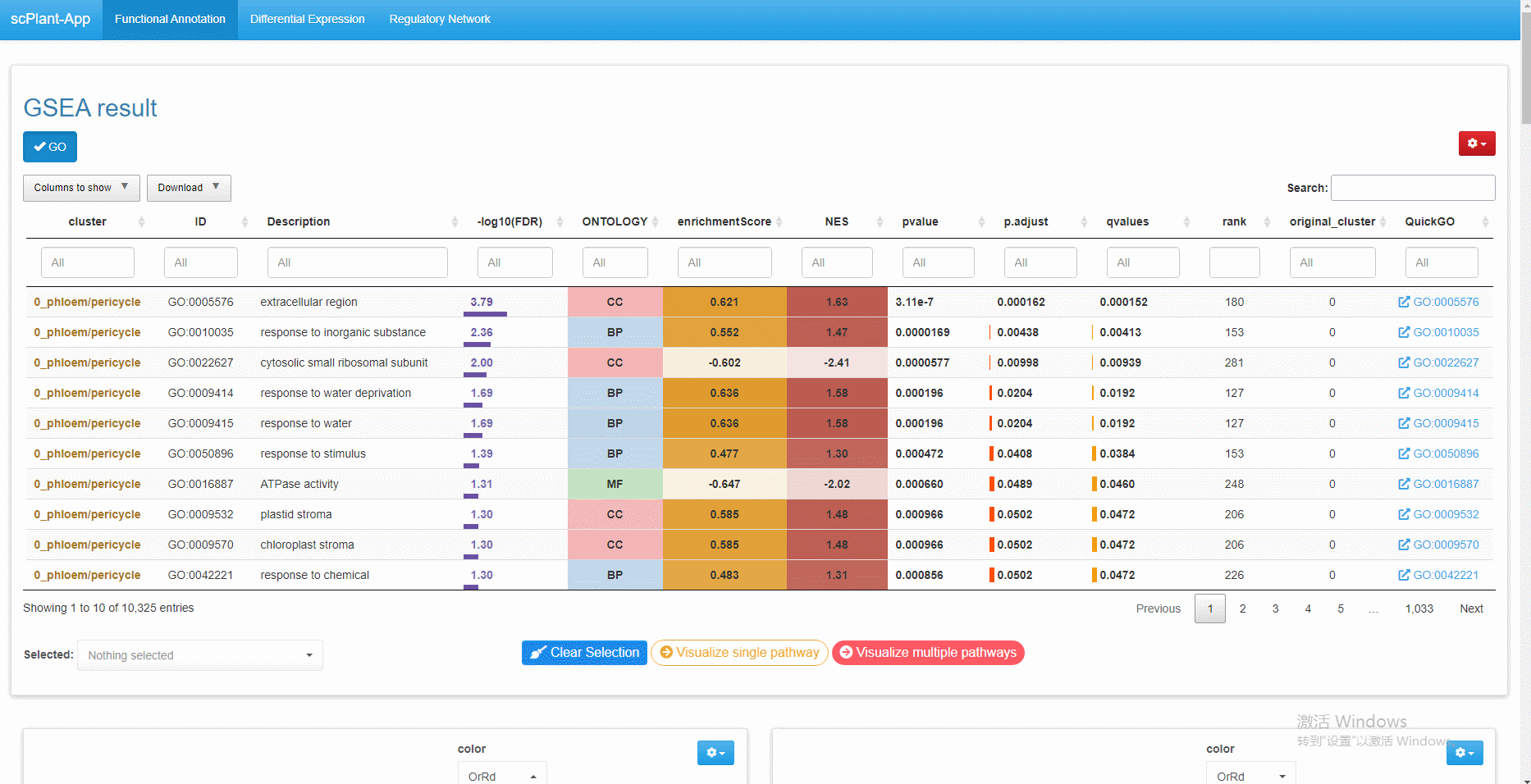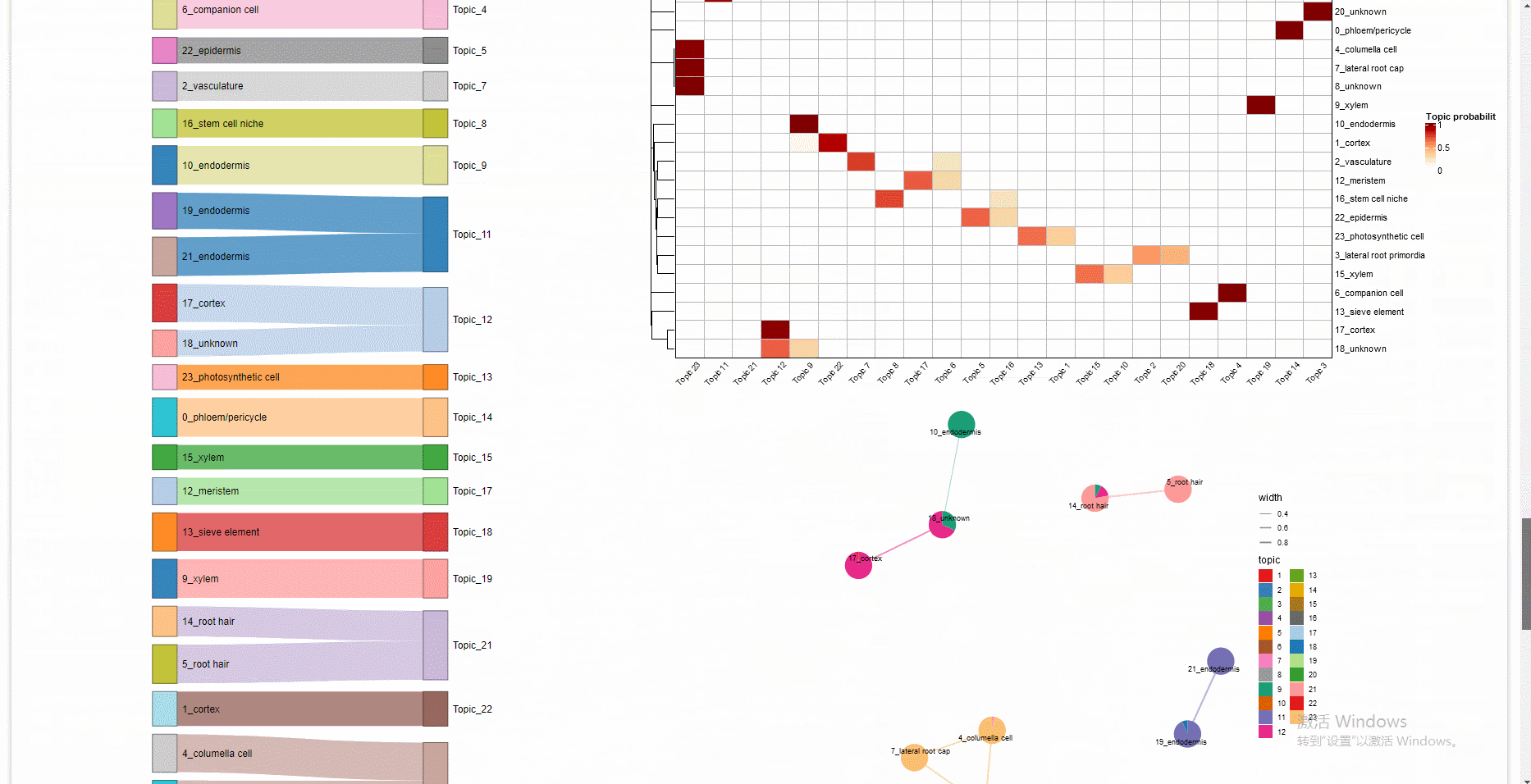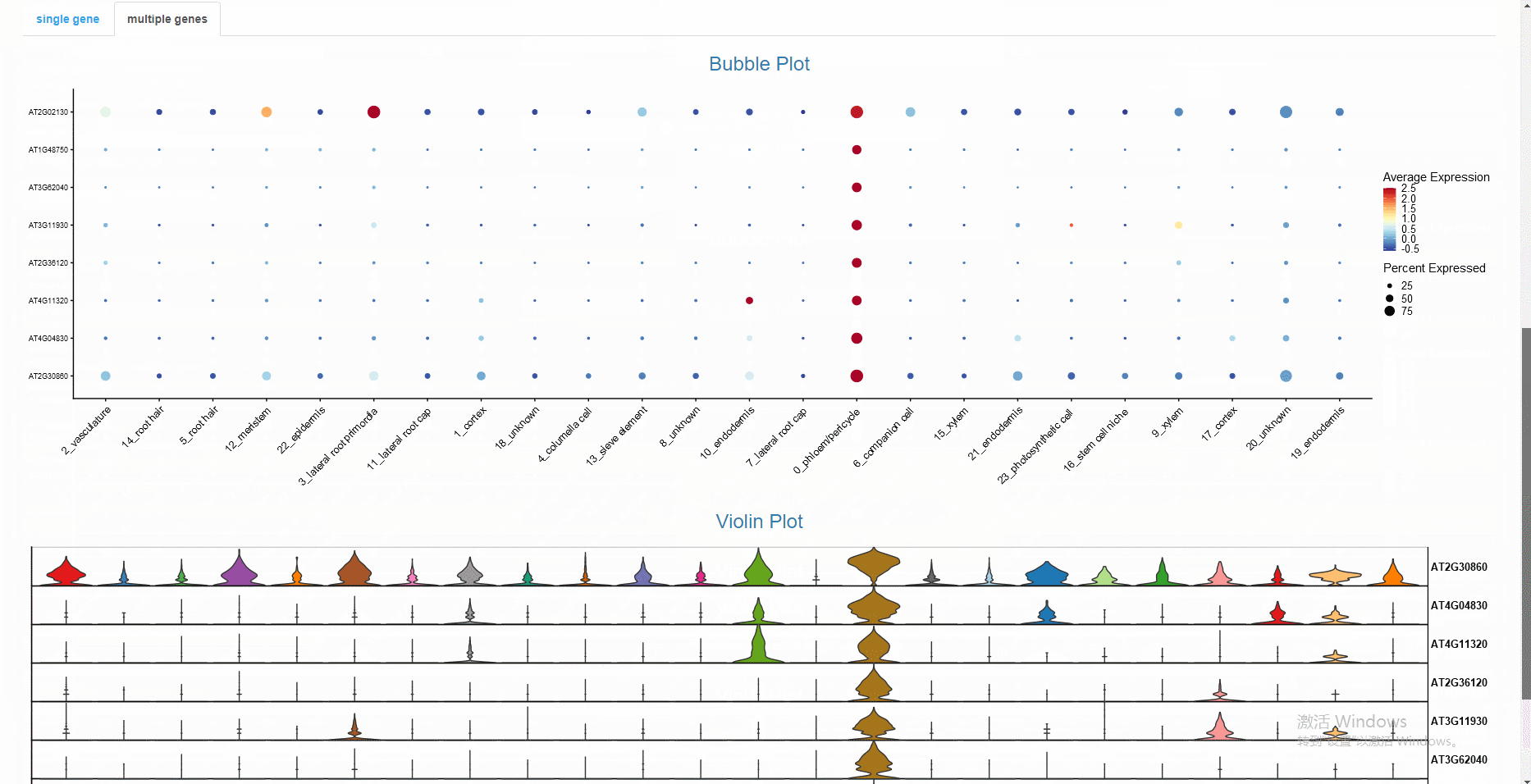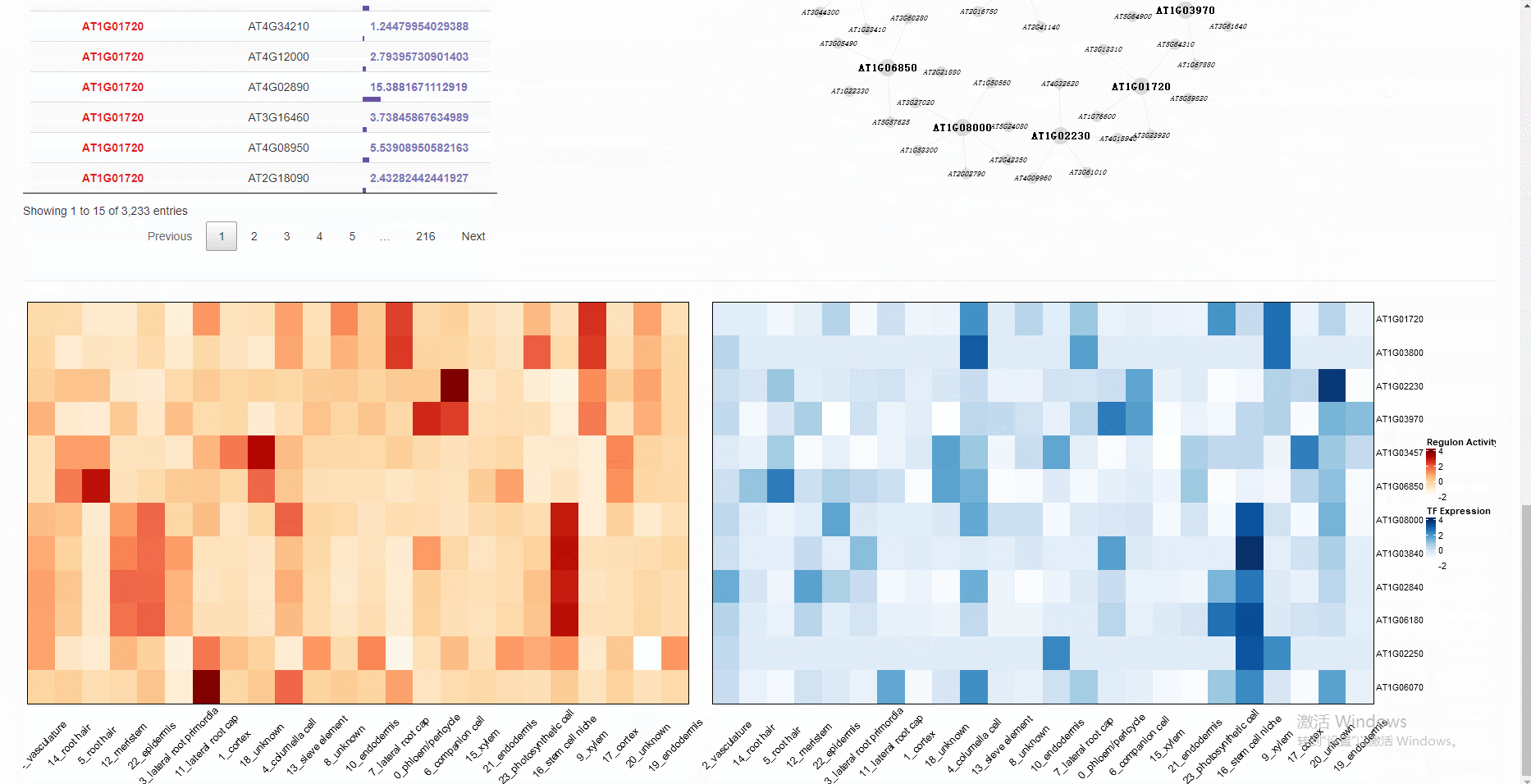
The gene regulatory network, differentially expressed genes, and functional annotation can be explored interactively in the built-in Shiny web application.
We provide a real scRNA-seq data of Arabidopsis thaliana (Zhang et al., 2019) as example data to show how to explore in built-in Shiny APP, download here
unzip exampleData.zip
SeuratObj <- readRDS("exampleData/SeuratObj.rds")
rssMat <- readRDS("exampleData/rssMat.rds")
rasMat <- readRDS("exampleData/rasMat.rds")
tf_target <- readRDS("exampleData/tf_target.rds")
library(scPlant)
load_shinyApp(SeuratObj, rasMat, rssMat, tf_target)
# Note that objects named "SeuratObj", "rasMat", "rssMat", "tf_target" are needed in your global environment.The Shiny app is demonstrated in the following image: